7.Error Code¶
7-1.Summary¶
| Error ID | Description |
|---|---|
| Err001 | Octopus-toolkit cannot access the web page. |
| Err002 | Incorrect GEO accession number. |
| Err003 | The experiment type cannot be handled with Octopus-toolkit. |
| Err004 | The data cannot be processed. |
| Err005 | Not enough disk space. |
| Err006 | Related to each processing step. |
| Err007 | Some analytics tools are not installed. |
| Err008 | Incorrect password. |
| Err009 | Octopus-toolkit can’t read/write files from your computer. |
| Err010 | Incorrect number of Paired-End data. |
If you have any questions, Please contact us at Octopustoolkit@gmail.com
7-2.Detail¶
Err001¶
Octopus-toolkit attempts to access the NCBI server (National Center for Biotechnology Information) to obtain sample information.
If your network connection is unstable, or the NCBI server is tempararily unavailable, Octopus-toolkit cannot get information for GSE and/or GSM.
First, check the network connection of your computer. If it is ok, please check the NCBI and whether the server is operating normally.
If the above cannot solve the problem, the connection to the NCBI may be timed out due to unknown reasons. Please re-run Octopus-toolkit after some time (temporary phenomenon).
Err002¶
Octopus-toolkit obtains sample information from the GEO (gene expression omnibus) website.
GEO Accession NumberA GSExxx is a unique GEO accession number assigned to a study. A GSMxxx is a unique GEO accession number assigned to a sample. A single GSE (study) can have a number of GSM (samples).
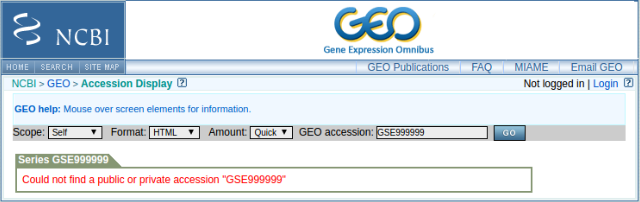
Octopus-toolkit can only process registered GSE or GSM ids in GEO. Err002 occurs when you put unregistered accession ids or misspelled accession ids.
- Unregistered GSE id (Input :
GSE999999)
- Misspelled or incorrect accession number (Input :
ChIP-Seq)
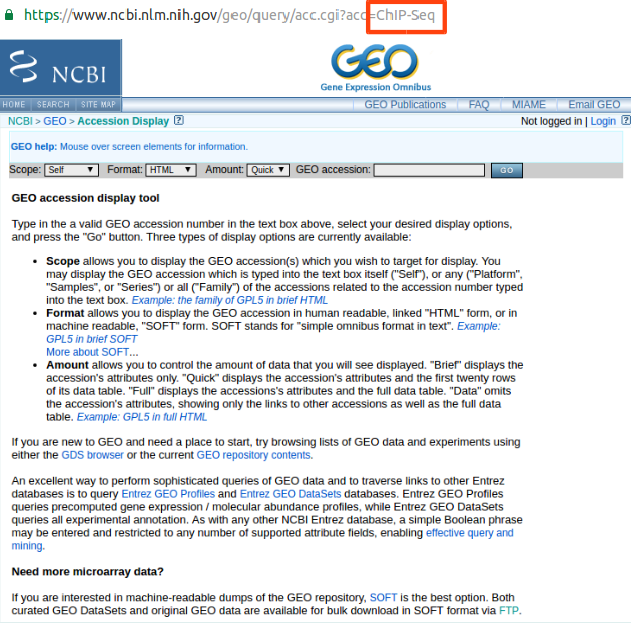
Please check the GEO accession number whether it is registered in the GEO.
Err003¶
There are many different types of next-generation sequencing (NGS) data. As defined by NCBI (NGS data - Study type), genome binding/occupancy profiling by high throughput sequencing indicates ChIP-seq data.
Octopus-toolkit currently supports the following types of NGS data. Other NGS types will be skipped.
expression profiling by high throughput sequencing (RNA-seq)
genome binding/occupancy profiling by high throughput sequencing (ChIP-seq / MNase-seq / ATAC-seq / MeDIP-seq / DNase-seq)
(Other NGS types will be added later)
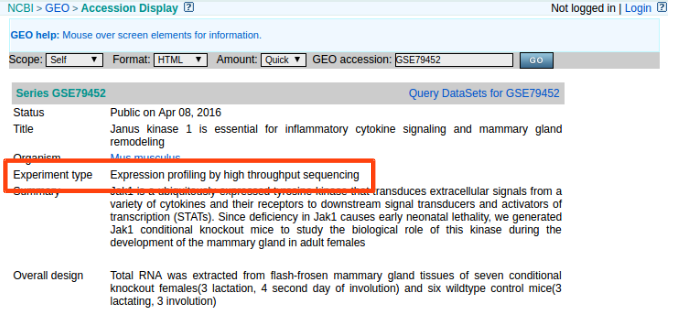
You can check experiment type of given GEO accession number through the website. (ex: GSE79452)
- Experiment Type
Err004¶
Not all data in the GEO can be processed with the Octopus-toolkit. Octopus-toolkit check the following information before the processing.
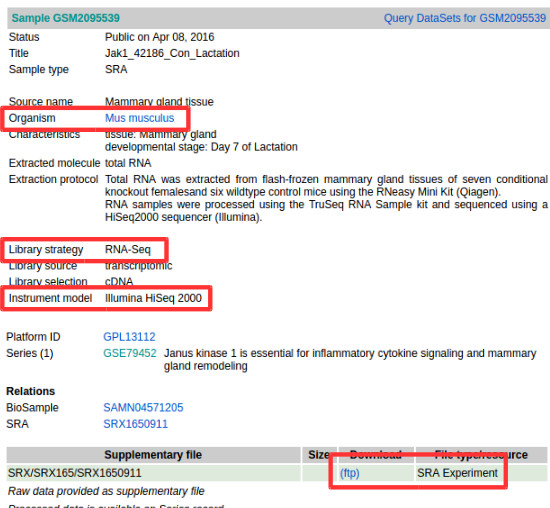
Organism, Library strategy, Instrument model, and FTP Address(SRA Experiment)). (Important)
- DataSet for GSE79452 (Ex : GSE79452)
Err004 is divided into the following four subcategories.
| Sub Error ID | Description |
|---|---|
Err004-1 |
The organism is not supported. |
Err004-2 |
The experiment type is not supported (for example Exome-seq). |
Err004-3 |
The instrument is not supported. Octopus-toolkit can only process data generated by Illumina instrument. |
Err004-4 |
Raw data (.sra) is currently unavailable (probably newly registered data). |
Err004 is related to unsupported data by Octopus-toolkit. The following data is currently handled with Octopus-toolkit.
| Type | Description |
|---|---|
| Organism | Homo sapiens, Mus musculus, Drosophila melanogaster, Saccharomyces cerevisiae, Canis lupus familaris, Arabidopsis thaliana, Danio rerio, Caenorhabditis elegans |
| Library Strategy | ChIP-Seq, RNA-Seq, MeDIP-Seq, ATAC-Seq, DNase-Seq, MNase-Seq |
| Instrument Model | Illumina GA/HiSeq/MiSeq (Illumina) |
Err004-4 indicates that data has been registered in the GEO, but the raw data (.sra) has not been released yet. Therefore, please check the availability of raw files.
Error004-4example : GSM1675769
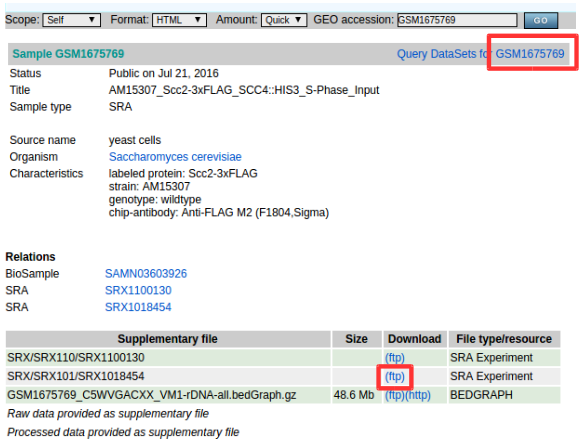
- No raw files (.sra).
Err005¶
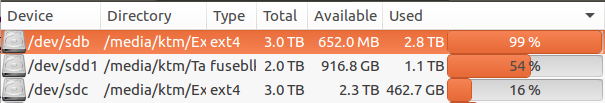
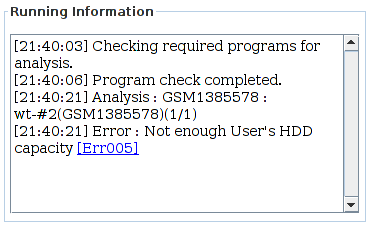
This error is related to disk space. To resolve this issue, obtain enough free space (more than 10Gb) and re-run the analysis.
- Check your hard disk space.
- Status window.
Err006¶
Err006 is divided into six subcategories.
| Sub Error ID | Description |
|---|---|
| Err006-1 | Cannot access NCBI’s FTP server. |
| Err006-2 | File converting error from .sra to .fastq using fastq-dump. |
| Err006-3 | Related to the .fastq file while checking the quality using FastQC. |
| Err006-4 | No input file (.fastq) for Trimming. |
| Err006-5 | Related to the Mapping step. |
| Err006-6 | Related to the Sorting step (BAM file). |
Err006-1¶
NCBI provides raw data of published sample through FTP server to user. If the NCBI homepage is working normally, you can extract the sample information, but if the FTP server does not work, you will not be able to download the data.
To solve this issue, you connect directly to the FTP server of NCBI.
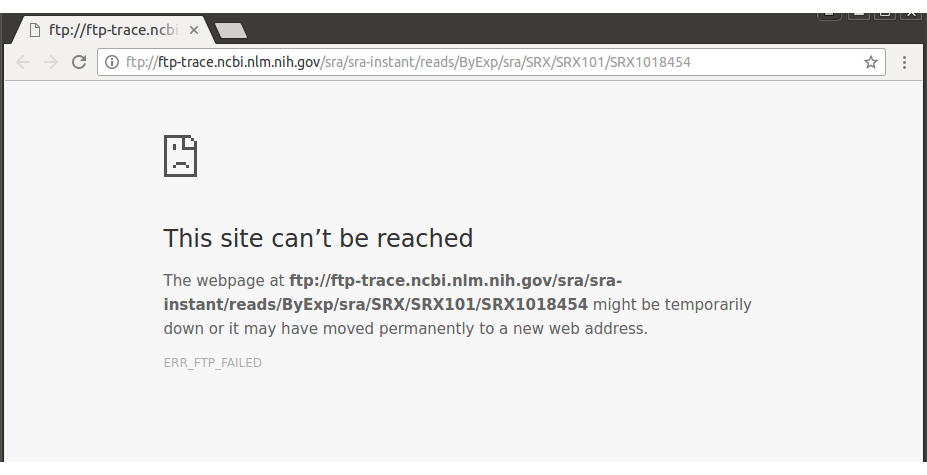
Error006-1example : GSM1675769
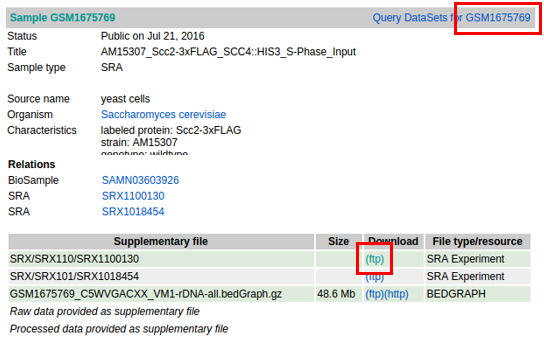
If you can connect to the FTP server, manually download the published sample.
- NCBI Ftp server is running.(
Success)
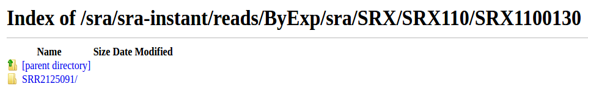
If the server is closed or samples can not be downloaded, please contact the NCBI because it is an issue for the NCBI.
- NCBI Ftp server is closed.(
Fail)
If the above method works normally, please try Octopus-toolkit again.
If you still have an Err006-1 in the retrial, please contact us at the address below.
Contact us : Octopustoolkit@gmail.com
Err006-2¶
Raw data of samples downloaded from NCBI is compressed in SRA format. For NGS analysis, SRA file should be converted to Fastq format. The tool used in this step is Fastq-dump, a sub tool of SRA-Toolkit.
Input file: Sequence Read Archive (Extension :sra)Output file: Short read sequence. (Extension :fastq)
006-2 occurs when there is no or invalid SRA file, which is the input file for executing Fastq-dump.
This error may arise due to an abrupt disconnection during the previous downloading step of the raw data from FTP server, or raw data uploaded to NCBI may be broken.
You should check your network status, free space on your computer and try the analysis again.
If the above method does not work, please contact us at the address below.
Contact us : Octopustoolkit@gmail.com
Err006-3¶
Err006-3 means that the input file(Fastq) for the Quality Check is invalid or an issue in the system itself during Quality Check using FastQC.
You should check fastq files on your computer and try the analysis again.
If the above method does not work, please contact us at the address below.
Contact us : Octopustoolkit@gmail.com
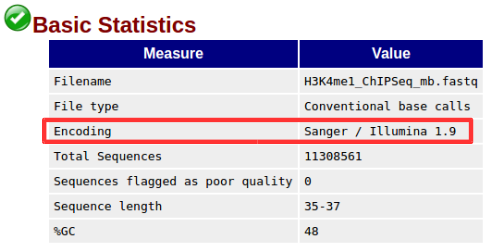
After successfully completing the Quality Check step, some problems may prevent FastQC from generating Fastqc_data.txt.
Octopus-toolkit extracts the encoding information of the sample from fastqc_data.txt among the outputs of FastQC. Therefore, if Fastqc_data.txt is not generated, it stores the encoding information of the latest samples. (Sanger / Illumina 1.9)
Err006-3Encoding information:
Err006-4¶
Err006-4 occurs when there is no input file(Fastq) for Trimming step or when all reads are removed due to bad quality.
You should check fastq files on your computer and try the analysis again.
If the above method does not work, please contact us using address below.
Contact us : Octopustoolkit@gmail.com
If all reads are removed by bad quality, Octopus-toolkit will use the non-trimmed input file(Fastq) to proceed. (Next step : Mapping)
Err006-5¶
Err006-5 may arise due to the following reasons.
- The input file (
non_trimmed Fastq,Trimmed Fastq) does not exist. - A large number of reads are trimmed due to
bad sequencing qualityorhigh threshold used. - Too few mapped reads (Less than 2 MegaByte).
You should check your input file (non-trimmed and trimmed fastq files), read count, file size after timming.
Err006-6¶
Err006-6: BAM (mapped) file does not exist or the number of mapped reads is too small.
You should check input file and BAM file.
Err007¶
Err007 is related to the installation step.
To use the Octopus-toolkit, your must follow the installation procedure completely: Requirement(Err007-1) and analysis tools(Err007-2).
- Requirement : Library files must be installed.
- Analysis tools : Tools are installed automatically by Octopus-toolkit. If the installation procedure is interrupted, please remove the Octopus-toolkit directory and rerun it.
Octopus-toolkit download files from the HOMER website. Err007 occurs if the website (http://homer.ucsd.edu/homer/) is unavailable, Err007 can occur.
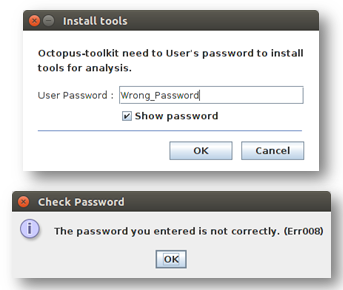
Err008¶
Err008 is related to password issue.
Password: You must enter your password once during the installtion step.
Please check your password and try again.
- When you enter incorrect password (Example : My password = ktm123)
Err009¶
Err009 is related to script files generated by Octopus-toolkit. If this happens, please rerun it later.
Err010¶
Err010 indicates that the number of files (paired-end sample) does not match when merging.
If there are several SRA files in one sample (GSM), Octopus-toolkit will merge them.
Paired-end data must have two files, Sample1_1.fastq and Sample1_2.fastq.
Err010 occurs if any of these fails.